Differantial interactions
Introduction
Chicdiff is an R package complementary to CHiCAGO, with the goal of identifying differential Capture Hi-C interactions. This can be very useful for comparing interactions found under different experimental conditions.
For detailed information on the package and installation, please see Chicdiff Github page. Here is a brief step by step installation (requires R version >= 3.4.0)
install.packages("devtools")
library(devtools)
install_github("RegulatoryGenomicsGroup/chicdiff", subdir="Chicdiff")
Input files
Peak files
Chicdiff uses a peak format of the interactions, based on the CHiCAGO output
Generating peak files:
First, generate a tab delimited list of all the samples and their associate Rds files (as generated under data directory from a CHiCAGO run), for which you want to find differential interactions. Make sure that all the datasets that you use were generated with the same Design files.
For this demonstration we will compare interactions between NSC (two replicas) and iPSC (two replicas). This information should be recorded in a tab delmited file, as in this example file, named Rd_calls.txt with the following content:
Rd_calls.txt:
NSC_rep1 /home/user/NSC_rep1_chicago_calls/data/NSC_rep1_chicago_calls.Rds
NSC_rep2 /home/user/NSC_rep2_chicago_calls/data/NSC_rep2_chicago_calls.Rds
iPSC_rep1 /home/user/iPSC_rep1_chicago_calls/data/iPSC_rep1_chicago_calls.Rds
iPSC_rep2 /home/user/iPSC_rep2_chicago_calls/data/iPSC_rep2_chicago_calls.Rds
The makePeakMatrix.R script generate a peak database based on the CHiCAGO databases that were detailed in the Rd_calls.txt file.
Rscript ./chicago/chicagoTools/makePeakMatrix.R --twopass --notrans --maxdist 3000000 Rd_calls.txt all_peaks
In this example peaks of cis interactions <3Mb are called.
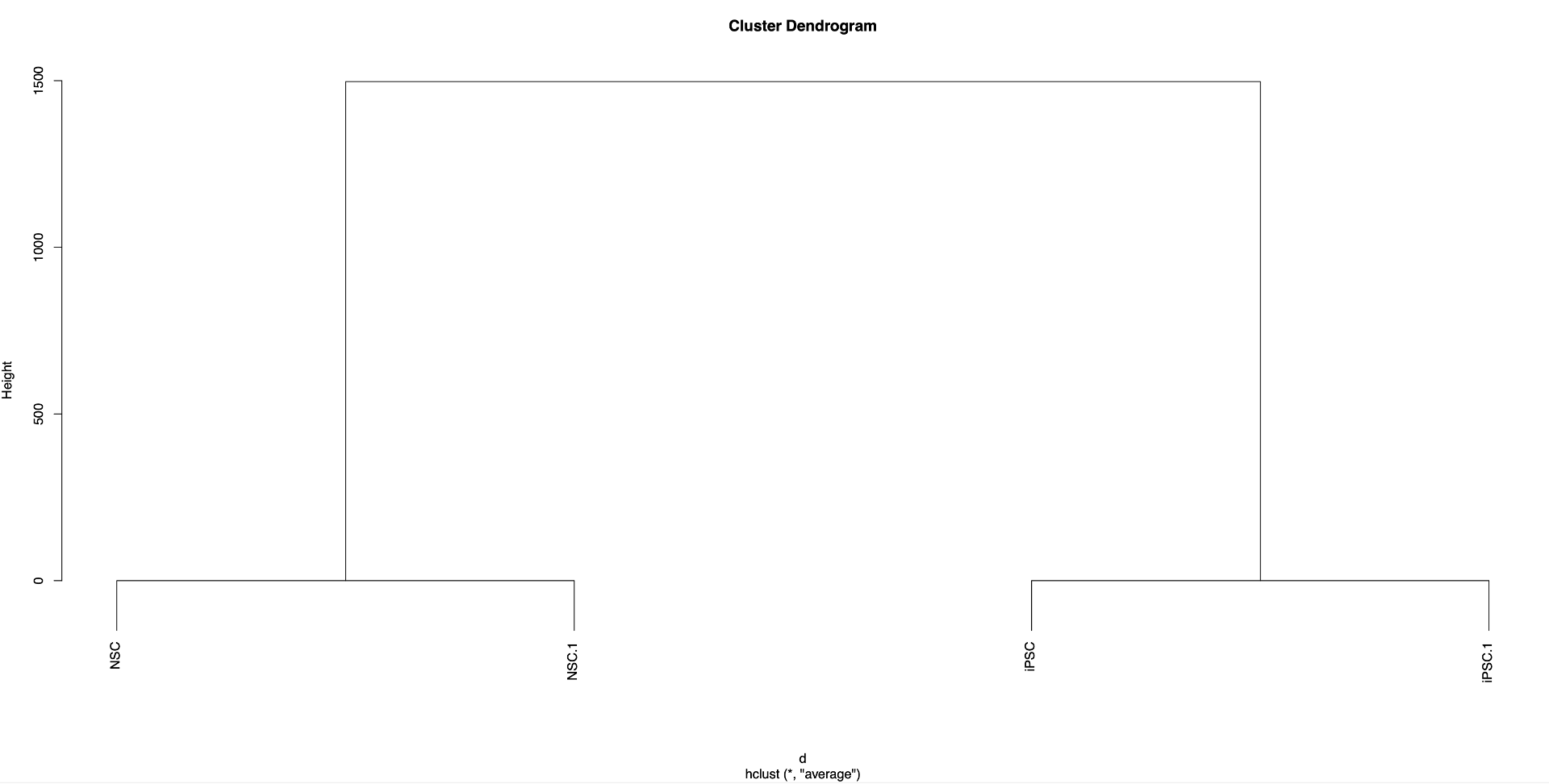
If multiple (>2) datasets were used in the makePeakMatrix.R step, a clustered dendrogram will be generated as well:
To analyze the data you will also need:
Design Library
Path to design library (same one as was used for generating the .Rds files above)
Chinputs
chinput files (same as the chinput files that were used for the CHiCAGO runs)
Calling differential interactions
In your R console, follow the principle of this example, with two replicas of NSCs and two replicas of iPSCs, make sure to be consistent with naming (e.g. to replica no. 1 of iPSC experiment use the same name for example iPSC_rep1 for all its associated files such as chinput, Rds etc’).
library(Chicdiff)
#set design library:
DesignDir <- file.path("/home/user/", "h_10kDesingFiles")
#set interaction data sets to be included:
ChicagoData <- list(iPSC_Micro = c(iPSC_rep1 = file.path("/home/user/10kb_run/iPSC_rep1/data","iPSC_rep1.Rds"), iPSC_rep2 = file.path("/home/user/10kb_run/iPSC_rep2/data","iPSC_rep2.Rds")), NSC_MicroC = c(NSC_rep1 = file.path("/home/user/10kb_run/NSC_rep1/data","NSC_rep1.Rds"), NSC_rep2 = file.path("/home/user/10kb_run/NSC_rep2/data","NSC_rep2.Rds")))
#set chinput files:
CountData <- list(iPSC_MicroC = c(iPSC_rep1 = file.path("/home/user/pooled_10kb_chinput/iPSC_rep1", "iPSC_rep1.chinput"), iPSC_rep2 = file.path("/home/user/pooled_10kb_chinput/iPSC_rep2", "iPSC_rep2.chinput")), NSC_MicroC = c(NSC_rep1 = file.path("/home/user/pooled_10kb_chinput/NSC_rep1", "NSC_rep1.chinput"), NSC_rep2 = file.path("/home/user/pooled_10kb_chinput/NSC_rep2", "NSC_rep2.chinput")))
#set peakfiles:
PeakFiles <- "/home/user/peaks/all_peaks.txt"
#set parameters for Chicdiff run:
chicdiff.settings <- setChicdiffExperiment(designDir = DesignDir, chicagoData = ChicagoData, countData = CountData, peakfiles = PeakFiles, outprefix="10kb", settings = list(parallel=TRUE))
#run Chicdiff
Diff <- chicdiffPipeline(chicdiff.settings)
#example, how to plot specific bait interactions using the bait number:
plotDiffBaits()
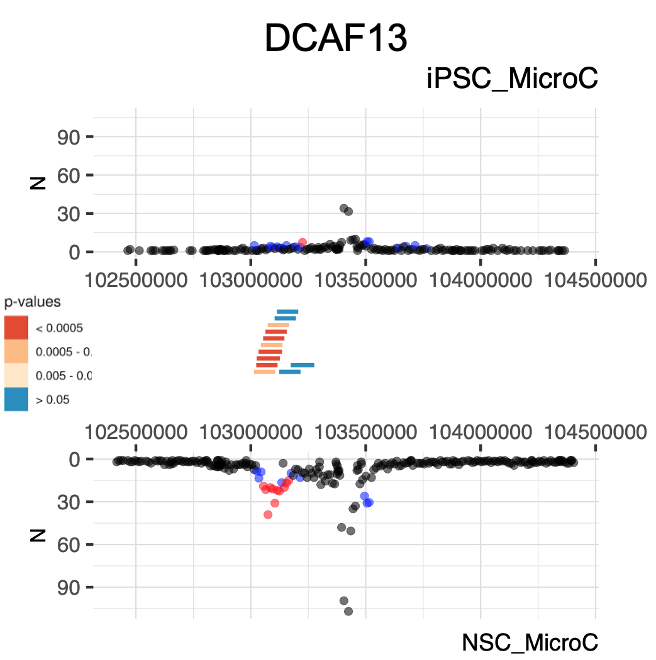
Additional information on how to use the package can be found on Chicdiff Vignette