Welcome to Dovetail Genomics® Promoter Panel data analysis documentation!
Overview
The Dovetail® Pan Promoter Enrichment Panels were designed to include all known coding genes and lncRNA promoters in the human and mouse genomes, respectively. The panels are to be used in conjugation with either Dovetail Omni-C® or Dovetail Micro-C® libraries for the detection of long-range interactions between gene promoters and their various distal-regulatory elements.
Spatial Control of Transcriptional Regulation:
Transcriptional regulation relies on interactions between protein complexes bound at the transcriptional start site (TSS) and at regulatory sites, including enhancers.
Promoter and enhancer sites can be separated by kilobases (or even megabases) of intervening sequence. Understanding these interactions is key to unravelling gene regulation and related human disease.
Promoter-enhancer (P-E) interactions cannot be detected by standard NGS approaches. For example, transcription factor binding to promoters or enhancers sequences is detectable using ChIP-seq maps protein binding to promoter or enhancer this approach is blind to any higher order P-E interactions that may be mediated by the bound transcription factors. Another established method, ATAC-Seq, provides information about nucleosome-free regions but does not offer information about P, E binding or P-E interactions. Lastly, Hi-C sequencing approaches can be used for chromatin loop calling but require a high sequencing depth and offer limited resolution, requiring about 1B reads for the human genome.
Key benefits of Dovetail® Pan Promoter Enrichment Panel:
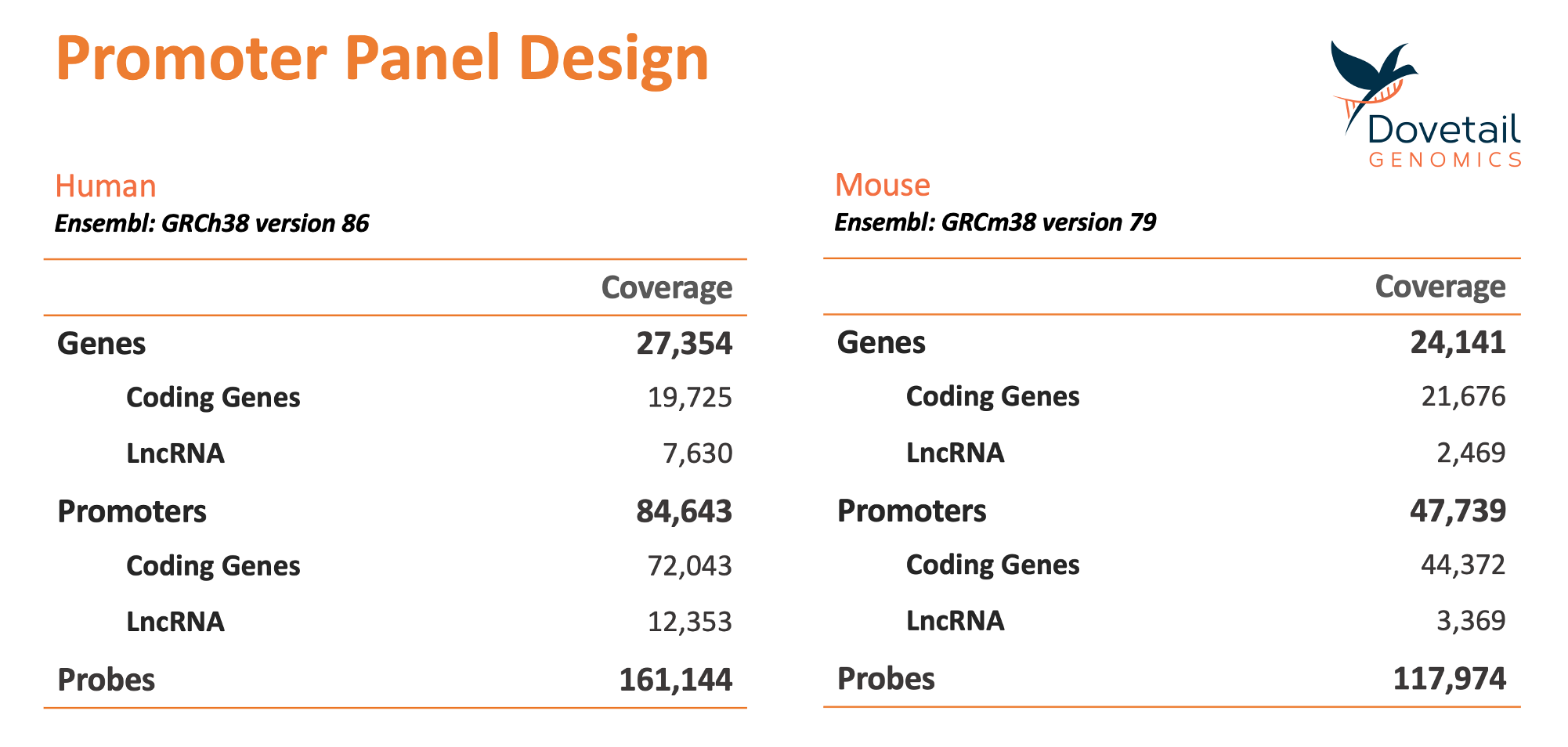
The panel was optimized to work using Dovetail’s unique restriction enzyme-free Hi-C, Omni-C and Micro-C technologies. Extemely high promoter coverage was achieved using this approach as no promoters were ommited from the panel due to lack of restriction sites in their vicinity as needed in other Capture Hi-C methods. The table below summarizes the genes, promoters and probes that are included in the human and mouse panels:
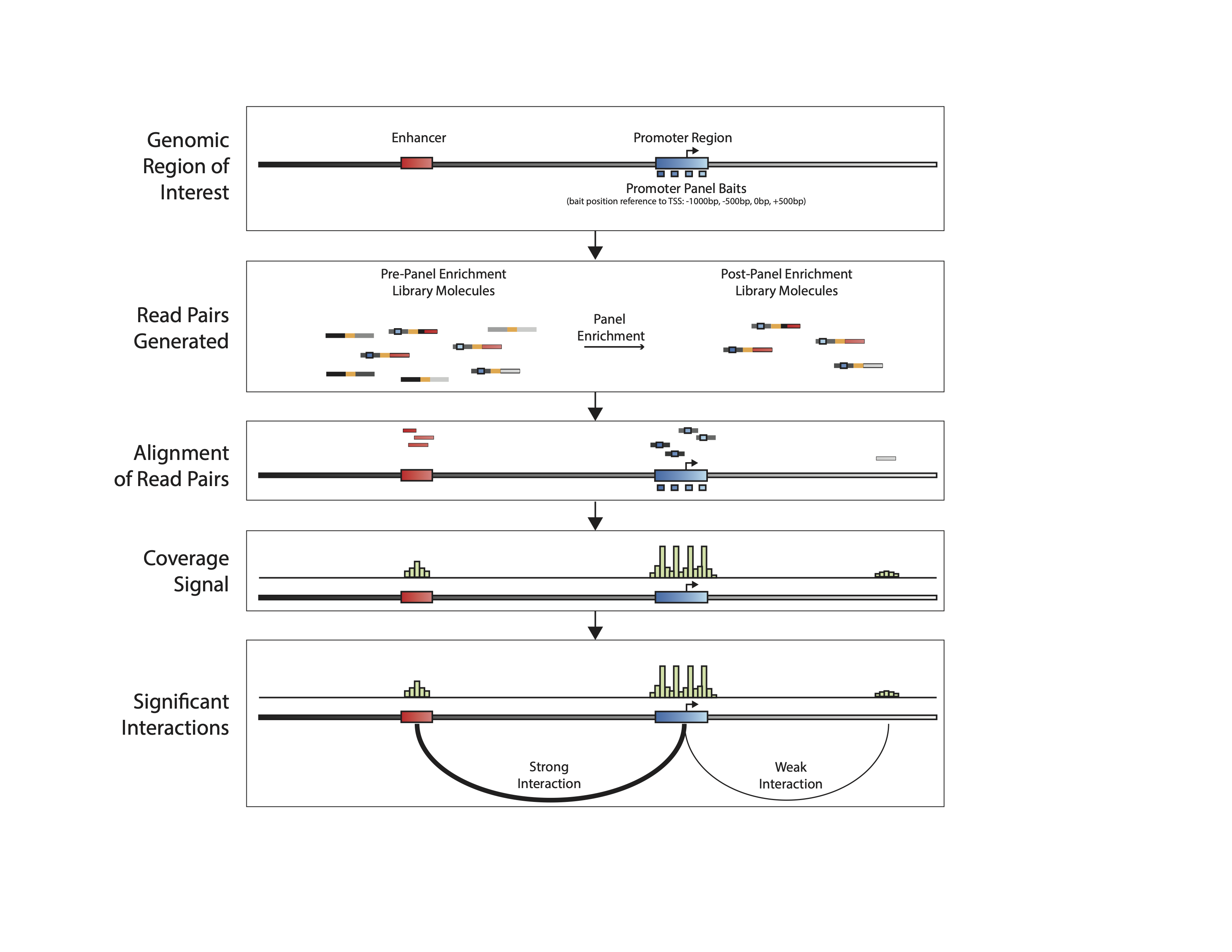
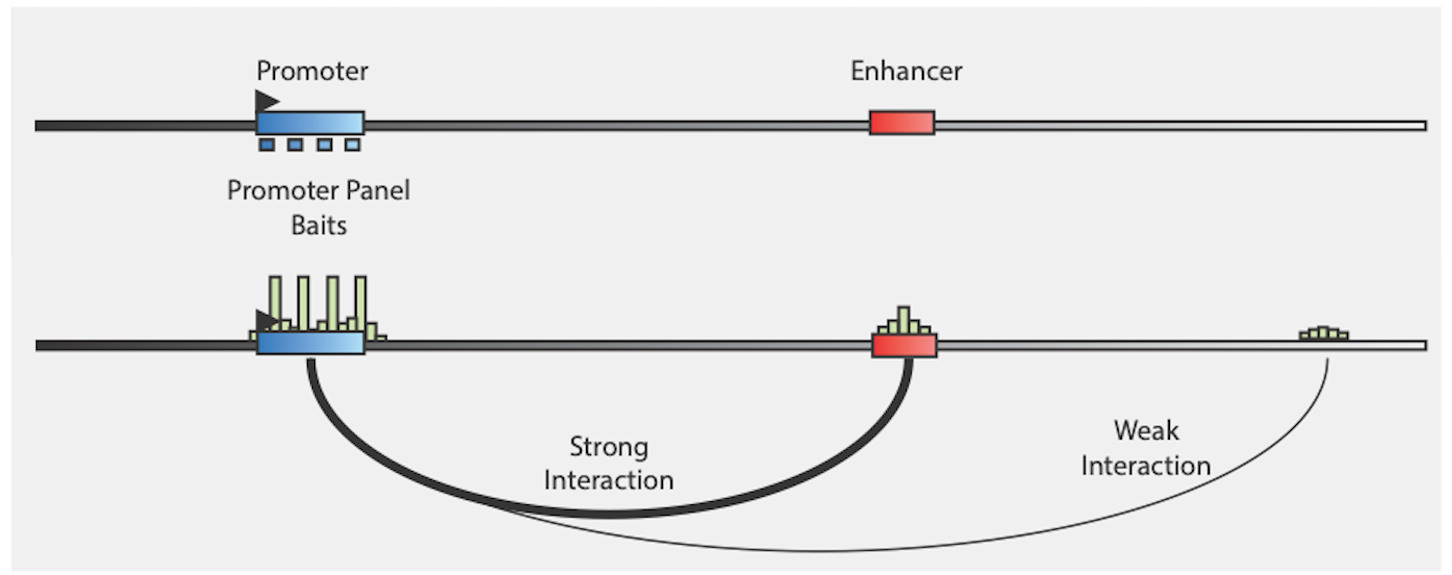
The Pan Promoter Enrichment Panel offers the highest resolution view of P-E interactions with lowest sequencing burden.
Captured Libraries maintain Hi-C characteristics in terms of valid read-pairs length distribution while achieving uniform coverage over baited regions.
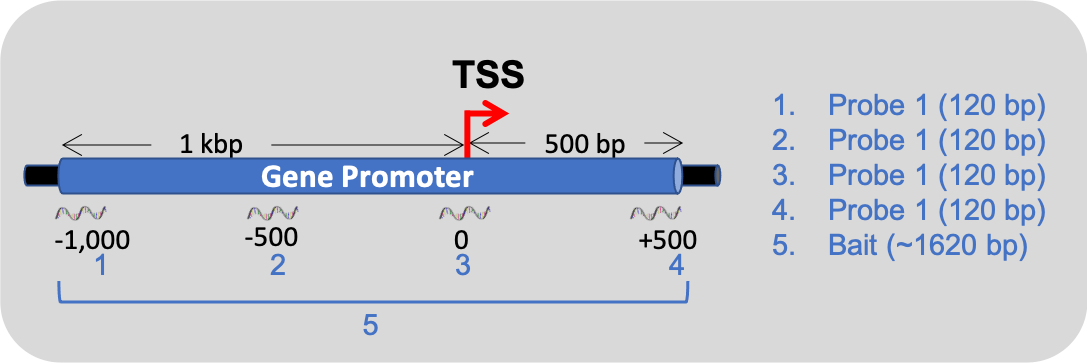
The reproducibility of the captured libraries is very high, with high correlation of coverage over baits between replicas, with typical \(R^2\) > 0.97. Baits are defined as the combined locations of probe targets that belong to same gene promoter, as area #5 in the image below:
This guide will take you step-by-step through the QC and interpretation of your captured library and evaluation of reproducibility between replicas. We will show you how to:
Generate contact maps
Find and prioritize significant interactions
Combine this information with additional data sets that you might have available.
Main steps in Pan Promoter captured library preparation and data processing:
If you have not yet sequenced a Dovetail captured library and simply want to get familiarize yourself with the data, an example dataset from iPSC and NSC captured Micro-C libraries can be downloaded from our publicly-available repository. In these data sets you will find bed files with the probes and bait locations.
If this is your first time following this tutorial, please check the Before you begin page first.
Contents: